Advisor Faculty Research Interests and Projects (P-Z)
Raddy Ramos, Ph.D., Associate Professor, Department of Biomedical Sciences

Cellular and synaptic changes of neurodevelopmental brain malformations
The overall goal of my research is to understanding how malformations of brain development lead to epilepsy and intellectual disability. Many parts of the brain such as the neocortex and cerebellum have elaborate cytoarchitecture which emerges from complex molecular/genetic programs during brain development. Not surprisingly, perturbations in these processes results in brain malformations which are accompanied by seizures and intellectual disability. Understanding how the brain circuits are rewired in the malformed brain is important toward developing new therapeutic strategies to help children with brain malformations.
MORE
Project 1: Cellular and synaptic changes in Gpr56 knockout mouse model of human brain malformation
GPR56 is gene linked to brain malformation in humans, characterized by abnormal lamination of the neocortex and cerebellum and accompanied by intellectual disability, language impairment, and epilepsy. Gpr56 knockout (KO) mice display similar brain malformations in the neocortex and cerebellum, suggesting that this mouse model has face validity as well as construct validity. Using in vitro and in vivo methods, we proposed to characterize the cellular and synaptic properties of neurons in the malformed Gpr56 KO brain and compare these observations with unaffected mice. A number of physiological tools will used including whole-cell patch-clamp recordings as well as whole-brain calcium imaging with genetically-encoded probes. We predict that neurons found in the wrong layers in Gpr56 KO mouse malformations will display hyper-excitability due to changes in their connectivity to other excitatory and inhibitory neurons. We propose to test how pharmacological manipulation of the GABAergic system can ameliorate these deficits.
Representative publications
Lipoff DM, Bhambri A, Fokas GJ, Sharma S, Gabel LA, Brumberg JC, Richfield EK, Ramos RL. 2011. Neocortical molecular layer heterotopia in substrains of C57BL/6 and C57BL/10 mice. Brain Res. 19;1391:36-43.
Mangaru Z, Salem E, Sherman M, Bhambri A, Brumberg JC, Richfield EK, Gabel LA, Ramos RL. Under review. Molecular Layer Heterotopia of Cerebellar Vermis: Cytoarchitecture and Transcriptional Profiles
Ramos RL, Smith PT, DeCola C, Tam D, Corzo O, Brumberg JC. 2008a. Cytoarchitecture and transcriptional profiles of neocortical malformations in inbred mice. Cereb Cortex. 18(11):2614-28.
Ramos RL, Tam D, Brumberg JC. 2008b. Physiology and morphology of callosal projection neurons in mouse. Neuroscience 153(3):654-63.
Olga V. Savinova, Ph.D., Assistant Professor, Department of Biomedical Sciences

Vascular calcification is an independent predictor of coronary artery disease and cardiovascular mortality. Using animal models, we demonstrated that overexpression of tissue-nonspecific alkaline phosphatase (TNAP) promotes vascular calcification and that animals with pre-existing TNAP-induced intimal calcification develop accelerated coronary artery disease. Our overall hypothesis is that under conditions that promote calcification (e.g. chronic kidney disease, diabetes, or advanced age) TNAP activity induces micro-calcification in the internal elastic lamina that accelerates retention atherogenic lipoproteins in the arterial wall. We also propose that inhibition of TNAP activity is a valuable therapeutic target for the reduction of atherosclerosis in patients predisposed to vascular calcification.
MORE
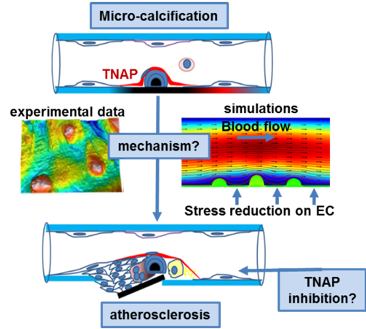
Aim 1. Mechanisms contributing to TNAP - driven acceleration of atherosclerosis
1.1. Cadaveric study (collaboration with Drs. Beatty and Plummer, NYIT COM)
We have recently published our findings demonstrating intimal and internal elastic lamina micro-calcifications in the coronary arteries from heart transplant patients. The prevalence of early calcific lesions will be further investigated in subjects older than 65. We will determine the distribution of calcium deposits in the tunica intima, media, and internal elastic lamina (IEL) in coronary, cerebral, and peripheral arteries.
1.2. Computational Fluid Dynamics simulations (collaboration with Dr. Beatty, NYIT COM, and Dr. Carka, NYIT CoECS)
Calcification can adversely affect luminal geometry and increase arterial stiffness. We will perform computational fluid dynamics (CFD) calculations in vessels modeled in silico based on the realistic representation of endothelial topology and vessel wall geometry in atherosclerotic mice with and without genetically engineered calcification.
Aim 2. Endothelial - specific ablation of TNAP in a model of chronic kidney disease (CKD)
2.1. A new mouse model (collaboration with Dr. Millan, SBP Institute, La Jolla, CA)
CKD is associated with increased vascular calcification and atherosclerosis. We are currently developing a new mouse model of tissue-specific TNAP deficiency. These mice will be used in a future study of CKD and will set the stage for pharmacological targeting of TNAP in CKD.
References
Romanelli F, Corbo A, Salehi M, Yadav MC, Salman S, Petrosian D, Rashidbaigi OJ, Chait J, Kuruvilla J, Plummer M, Radichev I, Margulies KB, Gerdes AM, Pinkerton AB, Millan JL, Savinov AY, Savinova OV. Overexpression of tissue-nonspecific alkaline phosphatase (TNAP) in endothelial cells accelerates coronary artery disease in a mouse model of familial hypercholesterolemia. PLoS One. 2017 12(10):e0186426
Savinov AY, Salehi M, Yadav MC, Radichev I, Millán JL, Savinova OV. Transgenic overexpression of tissue-nonspecific alkaline phosphatase (TNAP) in vascular endothelium results in generalized arterial calcification. J Am Heart Assoc. 2015 4(12). pii: e002499
ACTIVE funding
NIH R56HL131547 (PI), NSF DBI-1828305 (Co-PI)
Nikos Solounias, Ph.D., Professor, Department of Anatomy

My general goal is to understand the morphology and evolution of giraffes and bovids (antelopes). These ruminants are abundant and yet their evolution has not been studied in detail. They originate about 20 million years ago in Asia and Africa. I am especially interested in the evolution of horns and the limbs. I am also interested in the education of embryology using new methods.
MORE
Identifying giraffe species by osteologic differences. A few months ago, a geneticist in Frankfurt, Julian Fennessy, made a major discovery about giraffes. You may have read about this, it was all over the news- his team found that there are 4 separate species of giraffes. Their study was basically genetic and behavioral. Where they had data that certain individuals do not interbreed. Until now it was all thought the giraffe to be one species. Since this major article came out, Julian and I have been in contact about a potential collaboration. We will describe the skeletal morphology of these 4 species. This is a very important project. It will show osteologic differences. The findings can be extended to other groups and to fossils. We are now planning how to carry out this work- it will involve me going to collections in the United States and Europe to identify these species based on museum material, and describe the morphology of the four new species for the first time. I will therefore need time set aside to travel to study these specimens. I could write a grant proposal for this. The AMNH has at least two of these species (from Zaire and Kenya) and I have noted interesting differences on the bones. Such studies will be a major contribution to science and will also make our university known.
Siwalik giraffids and bovids; two anatomical monographs; well in the making. There is currently a large collection of Siwalik material housed at Harvard. I was part of the teams that excavated these in the 80s in Pakistan. We have these specimens on loan from Pakistan and are now housed at Harvard. The specimens will need to be returned to Pakistan in the next few years. There are thousands of specimens that no one has seen or published on. I am responsible for publishing on the giraffids and the bovids. Once they are returned to Pakistan, they will be inaccessible to westerners. Systematic works are being completed now. (1) Descriptions of the giraffe fossils are urgently needed. I can bring specimens here or I can work at Harvard with my student. I estimate 19 species were present which need to be described and published. Some new genera and species have been identified. (2) I am also involved in the bovid project (3,000 specimens), where we (Alan Gentry, John Barry and I) are describing and even naming some new genera and species. This project also has to be finished before the end of 2020 so it can be included in the book (as a summary).
Embryology 3D virtual textbook videos. I use clay to teach embryology. It is a popular style of lectures and students love it. We want to develop these lectures as virtual with 3D models to be developed. Such research and development would give the student a Ph.D in education.
Danowitz, M., Barry, J.C., Solounias, N. 2017. The earliest ossicone and post-cranial record of Giraffa. PLoS ONE 12:e0185139
Solounias, N., Danowitz, M. 2016. Astragalar morphology of selected Giraffidae. PLoS ONE 11(3): e0151310. doi: 10.1371/journal.pone.0151310
Rios-Ibanez, M., Danowitz, M., Solounias, N. 2016. First comprehensive morphological analysis on the metapodials of Giraffidae. Palaeontologia Electronica. 19:3.52A:1-39.
Randy Stout, Ph.D., Assistant Professor, Department of Biomedical Sciences

The roles and regulation of gap junction communication in the brain
The overall goal of my research is to understand how gap junctions contribute to brain function. I currently focus on the gap junctions that form intercellular and reflexive (autaptic) connections between/within astrocytes and oligodendrocytes. I use cell culture, mouse models, and computational simulations to test how changes (e.g. mutations) to the gap junction proteins (connexins) alter their behavior within cells and how such changes alter the interaction between connexins and other proteins.
MORE
I use microscopy and computational simulations to monitor the impact of experimental alterations to gap junctions and their interacting proteins on cell physiology and brain function.
Project Title: The Gap Junction Nexus
Specific Aim: Test the hypothesis that the supramolecular structure of gap junctions affects signaling between neural cells through actions of the carboxyl-terminus of the connexin proteins that make up the gap junction. (Actions of the C-terminus include posttranslational modifications that regulate channel opening, gap junction plaque structure, protein interactions with components of intracellular organelles, and gap junction protein turnover).
Approach: We will use three general techniques to most efficiently test a series of sub-hypotheses:
- Transgene expression in cultured cells with microscopy to study the spatial arrangement, mobility, and dynamic interactions of gap junction proteins with targeted mutations to experimentally probe the effects of block, or enhance the effects of protein domains in the C-terminus that regulate gap junction structure/function.
- Computational modelling to test how changes to gap junction structure/function will be predicted to recursively affect gap junction structure/function and to generate predictions on how dynamic changes in gap junction structure/function will alter intercellular signaling. Predictions based on modelling results will be tested using cultured cells with genetically encoded indicators for the intercellular signaling modality of interest (i.e. calcium indicators).
- Mouse models with altered gap junction proteins. We have Cx43 truncation and knockout lines currently breeding. New connexin mutant mouse models to target specific changes to gap junctions can be acquired in the future. We are now looking for changes in cell morphology and protein localization using immunofluorescence as a result of changes to gap junction protein mutations. We plan to extend this research in the next year to express connexins with targeted mutations and look for behavioral changes at the cell and whole organism level using calcium indicators and behavioral tests.
Published manuscripts directly related to this project:
Stout, Randy F., Erik Lee Snapp, and David C. Spray. "Connexin type and fluorescent protein-fusion tag determine structural stability of gap junction plaques." Journal of Biological Chemistry (2015): jbc-M115.
Stout, Randy F., and David C. Spray. "FRAP for the study of gap junction nexus macromolecular organization." Gap Junction Channels and Hemichannels. CRC Press, 2016. 63-91.
Stout Jr, Randy F., and David C. Spray. "Cysteine residues in the cytoplasmic carboxy terminus of connexins dictate gap junction plaque stability." Molecular biology of the cell 28.21 (2017): 2757-2764.
Aki Watanabe, Ph.D., Assistant Professor, Department of Anatomy
Research Overview: Developmental and Evolutionary Drivers of Phenotypic Diversity
My research seeks to answer one of the most enduring questions in biology—what drives phenotypic evolution? To this end, my work focuses on three areas: (1) the tempo and mode of morphological evolution at the micro- and macroscopic scale; (2) the complex interplay between anatomical changes along developmental and evolutionary time scales from days to 250 million years; and (3) the creation of new computational tools to investigate practical and theoretical issues in the collection and analysis of phenotypic data. These projects employ a synthesis of modern techniques, including high-resolution 3-D imaging, statistical shape analysis (geometric morphometrics), and programming. My previous projects have elucidated how the highly encephalized brains of birds evolved from their dinosaurian ancestors in a way that differs from how the mammalian and human brain evolved. In addition, I have developed new tools that allow users to evaluate the quality of their own morphological data. Beyond these studies, I regularly engage in paleontological fieldwork, including remote locations in Mongolia and Argentina. For additional and current information, please visit watanabe-research.com.
MORE
Project 1: Integration Pattern in Craniofacial Development and Evolution
I utilize geometric morphometric techniques to quantify and analyze changes and differences in shape of anatomical structures, particularly the skull and brain. As in most other craniofacial studies, my research to date have analyzed the skull and brain separately despite shared developmental pathways and physical interactions. To understand the complex interplay between these different tissue types, I am creating a 3-D atlas of the skull, brain, and muscles in developing alligators and chickens and identifying correlated shape changes (integration) among these tissues to see how these systems work together to form a cohesive, functional craniofacial anatomy.
Select publications
Watanabe, A., D.E. Slice. The utility of cranial ontogeny for phylogenetic inference: a case study in crocodylians using geometric morphometrics. Journal of Evolutionary Biology 27:1078–1092.
Watanabe, A., P.M. Gignac, A.M. Balanoff, T.L. Green, N.J. Kley, M.A. Norell. Are endocasts good proxies for brain size and shape in archosaurs throughout ontogeny? Journal of Anatomy. 10.1111/joa.12918
Project 2: Molecular Drivers of Brain Expansions
I am establishing a developmental biology lab at NYIT to greatly expand and complement my phenotypic work on vertebrate anatomy. Currently, I am interested in understanding the molecular mechanism underlying the highly encephalized brains of birds as a parallel system to brain enlargement in mammals and humans.
Project 3: Novel Analytical Method for Quantifying and Analyzing Protein Shape
As literal building blocks of life, proteins perform myriad functions critical for life. Their sheer diversity and functional specificity make them a suitable system to understand form-and-function relationships. For example, what changes to the protein structures enable certain animals to possess acute sense of smell, withstand potent toxins and venom, or survive in extremely cold environments? I have developed a novel computational tool to extract, quantify, and analyze shape information of proteins from online databanks. Preliminary results on the Trpc2 protein (Yohe et al. 2019) mathematically demonstrate that nectar-feeding bats with acute sense of smell exhibit a distinct shape in Trpc2 protein from other bats and mammals and that the major structural difference is highly localized to a single region on the protein. With this proof of concept, I am eager to extend this powerful framework to other proteins of enormous clinical importance, including hemoglobin, ribosome, rhodopsin, and venom proteins.
Select publications
Yohe, L.R., S. Hoffmann, A. Curtis. 2018. Vomeronasal and olfactory structures in bats revealed by diceCT clarify genetic evidence of function. Frontiers in Neuroanatomy 12:32.
Dong Zhang, Ph.D., Associate Professor, Department of Biomedical Sciences

The long-term goal of the Zhang lab is to understand the molecular mechanisms of how cancer cells respond to DNA replication stress. In addition, Zhang lab is investigating the feasibility of targeting the replication stress response pathway for cancer therapy.
Recently, Zhang lab discovered that FANCM (mutations of which leads to Fanconi Anemia syndrome), BLM (mutation of which leads to Bloom Syndrome), and BRCA1 (mutation of which leads to breast and ovarian cancers) collaboratively suppress replication stress response at the telomeres in cancer cells adopting the Alternative Lengthening of Telomere (ALT) as their telomere maintenance mechanism [1, 2]. In the same study, it was found that depletion of FANCM and BLM, or FANCM and BRCA1 using small interference RNA (siRNA) causes synthetic lethality in ALT cells. Synthetic lethality is defined as a scenario in which the simultaneous presence of two non-lethal mutations becomes lethal to a cell (Fig.1). Two new oncology drugs, LYNPARZA and RUBRACA, achieve their therapeutic efficacy by inducing synthetic lethality in ovarian cancers carrying BRCA1/2 mutations. Therefore, the discovery of new synthetic lethal interaction will uncover new oncology drug targets.
MORE
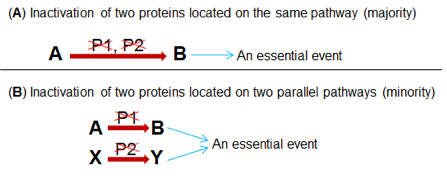
Built on this study, we recently performed a targeted screening and identified a cohort of factors that are implicated in the replication stress response at ALT telomeres. The DO/Ph.D. student will work in a collaborative team to achieve two goals: (1) Define the biological role of these factors in the replication stress response at ALT telomeres. (2) Explore the feasibility of targeting these factors for the treatment of ALT cancers.
References
- Pan, X., et al., FANCM, BRCA1, and BLM cooperatively resolve the replication stress at the ALT telomeres. Proc Natl Acad Sci U S A, 2017. 114(29): p. E5940-E5949.
- Pan, X., et al., Breaking the end: Target the replication stress response at the ALT telomeres for cancer therapy. Mol Cell Oncol, 2017. 4(6): p. e1360978.
Youhua Zhang, MD, Ph.D., Associate Professor, Department of Biomedical Sciences

Autonomic stimulation on atrial fibrillation in heart failure
The overall goal of my research is to understand better the role of the autonomic nervous system (sympathetic and vagal nerve) in the pathogenesis of atrial fibrillation (AF) and heart failure (HF). AF and HF are 2 common cardiac diseases, frequently coexist and can precipitate each other. The autonomic nervous system plays an important role in the pathogenesis in both diseases. Although the role of the autonomic nervous system in AF or in HF has been extensively studied, however, previous research has been focused on autonomic system in AF or in HF alone. Moreover, prior studies examining the effect of autonomic system on AF were typically conducted in normal animals, which are resistant to AF induction. Our recent data indicated that the response of failing hearts to autonomic stimulation is quite different from normal hearts. Failing hearts are more vulnerable to sympathetic stimulation (SS) induced AF and less sensitive to (strong) vagal stimulation (VS) induced AF. The mechanisms underlying the different responses of failing hearts versus normal hearts to autonomic stimulation induced AF remain to be studied.
MORE
Project 1: Define the role of cardiac ryanodine receptor (RyR2) dysfunction and increased Ca2+ leak as a potential mechanism leading to SS-enhanced AF arrhythmogenesis in HF.
It is known that RyR2 dysfunction exists in both AF and HF, and β-adrenergic stimulation could induce RyR2 dysfunction. We hypothesized that SS could exacerbate RyR2 dysfunction more easily in failing hearts than in normal hearts, contributing to SS-enhanced AF in HF. Experiments in this project will provide evidence from isolated myocytes, in vitro tissue preparations, to in vivo whole animals, that failing hearts have increased vulnerability to SS-induced RyR2 dysfunction and Ca2+ leak, leading to SS-enhanced arrhythmia inducibility. As a result, we expect that stabilizing RyR2 can ameliorate AF arrhythmogenesis in HF.
Selected publications
Delfiner MS, Nofi C, Li Y, Gerdes AM, Zhang Y. Failing Hearts Are More Vulnerable to Sympathetic, but Not Vagal Stimulation-Induced, Atrial Fibrillation-Ameliorated with Dantrolene Treatment. J Card Fail. 2018 Jul;24(7):460-469. PMID: 29885493
Project 2: Determine the role of VS on AF in HF.
We have demonstrated previously that VS induced AF is intensity-dependent, low-intensity VS is not arrhythmogenic on AF in normal animals. We have also demonstrated that chronic VS treatment can attenuate HF development. Since there is evidence of altered response of failing hearts to autonomic stimulation induced AF, experiments in this project will systematically investigate the different VS levels on AF in HF, and will delineate the potential mechanisms leading to the altered responses.
Selected publications
Zhang Y, Popovic ZB, Bibevski S, Fakhry I, Sica DA, Van Wagoner DR, Mazgalev TN. Chronic vagus nerve stimulation improves autonomic control and attenuates systemic inflammation and heart failure progression in a canine high-rate pacing model. Circ Heart Fail. 2009 Nov;2(6):692-9. PMID: 19919995
Zhang Y, Ilsar I, Sabbah HN, Ben David T, Mazgalev TN. Relationship between right cervical vagus nerve stimulation and atrial fibrillation inducibility: therapeutic intensities do not increase arrhythmogenesis. Heart Rhythm. 2009 Feb;6(2):244-50. PMID: 19187919
Haotian Zhao, Ph.D., Associate Professor, Department of Biomedical Sciences

Understanding of the Biology of Childhood Brain Cancers for Better Treatments
Brain tumors are the most common solid tumors in children and the most frequent cause of death from childhood cancers. In the era of precision medicine, choice of cancer treatment has been increasingly prescribed based on the molecular or genomic properties of the individual cancer. Indeed, the genomic and epigenetic landscapes of childhood brain cancers are vastly different their adult counterparts, indicating unique biology that require distinct therapeutic strategies. However, only a few new drugs have been approved that were specifically developed to treat children with cancer in the last 20 years. Children with brain cancers receive similar treatment regimens including surgery, radiation and chemotherapy used for adult brain cancers, leaving survivors with life-long debilitating complications from their treatments. Additionally, despite progress in the survival of children with brain cancers, there are still some brain cancers in children that are poorly understood, highly lethal with no effective treatment. Our laboratory is focused on how the growth of brain cells is regulated, and how its disruption contributes to brain cancers in children. This information will equip us to develop new therapies that specifically target tumor growth without damaging the developing brain. Our laboratory utilizes cellular, genetic approaches, and next-generation sequencing technologies to illustrate the dynamic transcriptomic and epigenetic landscapes in childhood brain tumors. We have developed multiple preclinical models with genetically engineered animals to understand the biology of pediatric brain tumors and to identify safer and more effective therapies.
MORE
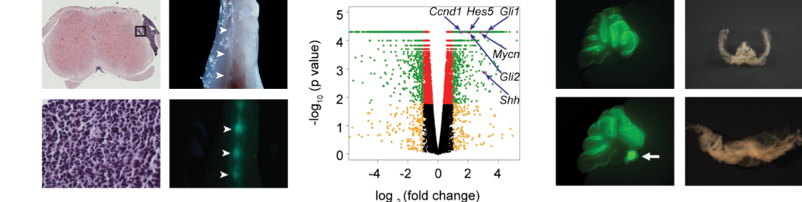
Project 1. Leptomeningeal metastasis.
Medulloblastoma (MB) is the most common malignant pediatric brain tumor that arises from the cerebellum. Leptomeningeal metastasis, frequently present at diagnosis and recurrence of MB, occurs when tumor cells spread to the leptomeninges along the surface of the brain and spinal cord. Recurrence/metastasis are serious complications with substantial morbidity and mortality. Our laboratory has generated animal models of MB with leptomeningeal metastasis that faithfully recapitulate the human disease. We will use these preclinical models to understand the molecular and cellular mechanisms of MB metastasis, and utilize the robust platform to evaluate therapeutic candidates to facilitate the development of effective new treatment for this highly lethal form of cancer metastasis.
Project 2. Rare pediatric brain cancer.
The choroid plexus (CP) in the brain ventricles is comprised of a fibro-vascular core lined by epithelial cells and responsible for the production of cerebrospinal fluid. CP tumors are rare primary brain tumors predominantly found in young children and infants. Though surgery offers excellent prognosis for the benign CP papilloma, few treatment options are available for the poorly understood and highly lethal CP carcinoma. We developed animal models that develop CP papilloma driven by sustained NOTCH1 signaling, whereas constitutive activation of both NOTCH and Sonic Hedgehog pathways leads to aggressive CP tumor that resembles CP carcinoma. Unlike CP epithelial cells which possess multiple primary cilia, NOTCH-activated CP tumor cells exhibit a solitary primary cilium. Conversely, most human CP tumors, especially CP carcinomas, comprise mono-ciliated cells characterized by suppression of the multi-ciliation program, indicating the importance of a singular primary cilium in CP tumor. In this project, we will develop an animal model using orthotopic transplantation of mouse CP carcinoma cells. To determine whether the multi-ciliogenesis program represents a potential therapeutic vulnerability in CP tumor, we will use a novel NOTCH inhibitor to determine whether NOTCH inhibition induces multiple cilia and suppresses CP carcinoma growth. Successful completion of these studies will help to translate the novel findings into improved outcome for patients with CP carcinoma.
Yingtao "Jerry" Zhao, Ph.D., Assistant Professor, Department of Biomedical Sciences

Genomics, epigenetics, computational biology, and Machine Learning
My research leverages multi-omics and computational approaches to understand the molecular basis of the epigenome, particularly in the healthy and diseased brain.
Project 1: Understanding the transcription and co-transcriptional processing of long genes
The lengths of protein-coding genes in the human genome range from 0.2 kilobases (kb) to 2400 kb. Long genes consume more time and resources to make a transcript because gene transcription relies on RNA polymerase II traveling along the entire gene region. Notably, misregulation of long gene expression has been linked to multiple human diseases, including amyotrophic lateral sclerosis, autism spectrum disorder, Alzheimer's disease, and cancer. We hypothesize that long genes require specific chromatin features to overcome the length constraint for efficient transcription and co-transcriptional processing. Consistent with this hypothesis, we found that a group of neuronal long genes harbor broad enhancer-like chromatin domains in their gene bodies. We will utilize functional genomics and computational approaches to investigate neuronal long genes.
MORE
Selected publications
- Ying-Tao Zhao, Deborah Kwon, Brian Johnson, Maria Fasolino, Janine Lamonica, Yoon Jung Kim, Boxuan Simen Zhao, Chuan He, Golnaz Vahedi, Tae Hoon Kim, Zhaolan Zhou. (2018) Long genes linked to autism spectrum disorders harbor broad enhancer-like chromatin domains. Genome research.
- Ying-Tao Zhao, Maria Fasolino, Zhaolan Zhou. (2016) Locus- and cell type-specific epigenetic switching during cellular differentiation in mammals. Frontiers in Biology.
Project 2: Elucidate the impact of the epigenome on the healthy and diseased brain.
Neurons carry unique epigenomic features compared to other somatic cells, and epigenomic deregulation is involved in the etiology of many brain disorders such as autism spectrum disorders (ASDs) and primary brain tumors. Indeed, genetic variants associated with ASDs are enriched in genes encoding chromatin regulators. Previously, I found that the expression of a group of neuronal genes is vulnerable to mutations in ASD-associated chromatin regulators and to high grade glioma. We plan to utilize multi-omics approaches, such as RNA-seq, GRO-seq, ChIP-seq, ATAC-seq, Hi-C, single cell omics, and Machine Learning, to elucidate the impact of the epigenome on the healthy and diseased brain.
Selected publications
- Ying-Tao Zhao, Darren Goffin, Brian S. Johnson*, Zhaolan Zhou. (2013) Loss of MeCP2 function is associated with distinct gene expression changes in the striatum. Neurobiology of Disease.
- Brian S Johnson, Ying-Tao Zhao, Maria Fasolino*, Janine Lamonica, Yoon Jung Kim, George Georgakilas, Kathleen Wood, Daniel Bu, Yue Cui, Darren Goffin, Golnaz Vahedi, Tae Hoon Kim , Zhaolan Zhou. (2017) Biotin tagging of MeCP2 in mice reveals contextual insights into the Rett syndrome transcriptome. Nature Medicine.
- Deborah Y Kwon, Ying-Tao Zhao, Janine M Lamonica, Zhaolan Zhou. (2017) Locus-specific histone deacetylation using a synthetic CRISPR-Cas9-based HDAC. Nature Communications.
- Yao Yao, Paul J Minor, Ying-Tao Zhao, … , Douglas Epstein. (2016) Cis-regulatory architecture of a brain signaling center predates the origin of chordates. Nature Genetics.